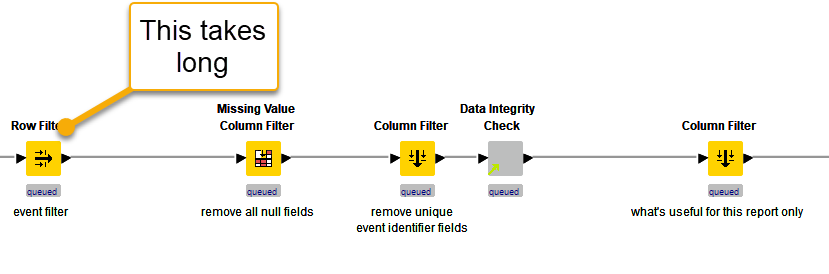
@Gavin_Attard
Couple of questions not yet asked:
- What version of KNIME are you using?
- Are you using the Apache Parquet columnar tables backend? Preferences->KNIME->Table Backend->Default (you may need to install Columnar Table Backend extension).
- How much memory have you allocated?
- How complex are the items you are filtering on - if strings, how long are they and is there a high degree of correlation in the first couple of letters?)
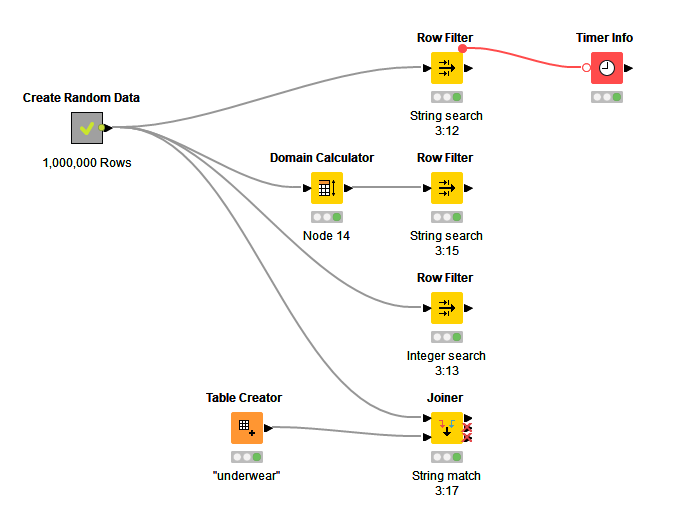
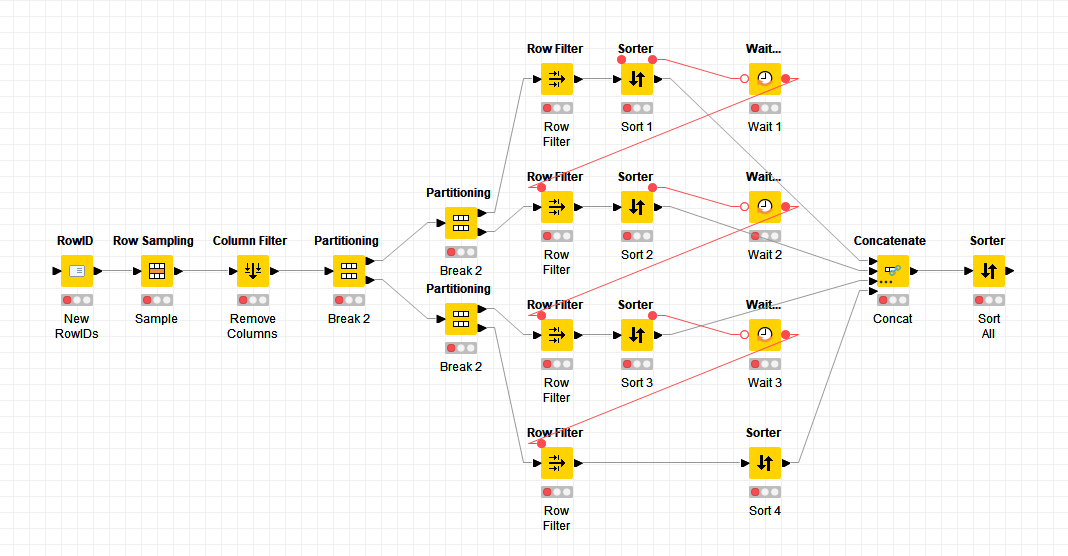
I’ve created a sample workflow (Scalable Row Filter 2023-07-02 PA1 – KNIME Community Hub) with 1,000,000 rows and average time to filter the data with 80 categories is less than 500mSec. So, if your workflow is taking a long time there is scope to optimise.
String matching
The first consideration is the nature of the data that you are matching. Consider the following strings:
“PROD-001-456”
“PROD-001-457”
When matching strings, the computer will compare the first letter, the second, etc…until it either reaches the end of the string and confirms the strings match, or finds a letter that doesn’t match and aborts early. Therefore, if your strings are long matching is going to take a long time and row filter will have a correspondingly long execution time. Also, if the first parts of the string are common across all the strings you will introduce a constant long delay in the matching process as each common letter is tested, until letters which identify a difference are reached.
Can you remove redundancy in the data by trimming strings to remove redundant (common) information? i.e. matching a substring (e.g. last four characters) rather than all characters.
The second optimisation is to replace the string with an integer (index number) that represents the category. Matching integers is a fast process, is a native function within the CPU, and is what most database engineers would do (normalise the data so that the main (fact) table contains indexes that point to dimension tables that contain the category descriptions). It is far quicker to match on integer indices, improves data processing speeds and reduces memory usage.
[Note, the table backend uses this approach with string columns - it creates a dimension table to store common strings, and then a fact table containing rows with pointers to the actual strings. By reducing redundancy it can significantly reduce the size of data stored to disc.]
Can you add an index column (integer) that represents the string to be match, then filter on the integer rather than the string?
The workflow that I created considered four ways of filtering the rows:
- Filtering on strings, which took 531mSec
- Filtering on integers, which took 339mSec - note, if you don’t have a column with integers you would need to factor in the time take to create a column. Easily done by grouping the original data column to get a list of categories, adding on integer sequence using the Counter Generation in the modular data generator extension then joining this integer index back to the original table using a joiner node.
- I’m not 100% sure how the row filter node works - I am making a guess that if there is domain information that says there are a limited number of strings (e.g. it is categorical data not random text), then it will use the category indices to do the match, not the actual strings. KNIME domain assumes a column is categorical if there are less than 60 different permutations of strings in a column, however, this limit can be raised using the Domain Calculator by unchecking Restrict The Number of Possible Values. This reduced the row filtering time to 391 mSec, however, the Domain Calculator took 607mSec. If you have long strings then this may give you a positive benefit - I don’t know your data, so don’t know.
- You can filter using a join node. This took 361mSec on string matching. Sometime, this approach works better than row filter node.
As always, your results may very and a lot will depend upon your data. If you are not using the columnar backend you will see a big improvement in using it. Otherwise, if you are matching strings then you can either manipulate the strings to improve matching performance or replace them with an index.
Hope that helps
DiaAzul
LinkedIn | Medium | GitHub